Gene essentiality is a quantitative trait
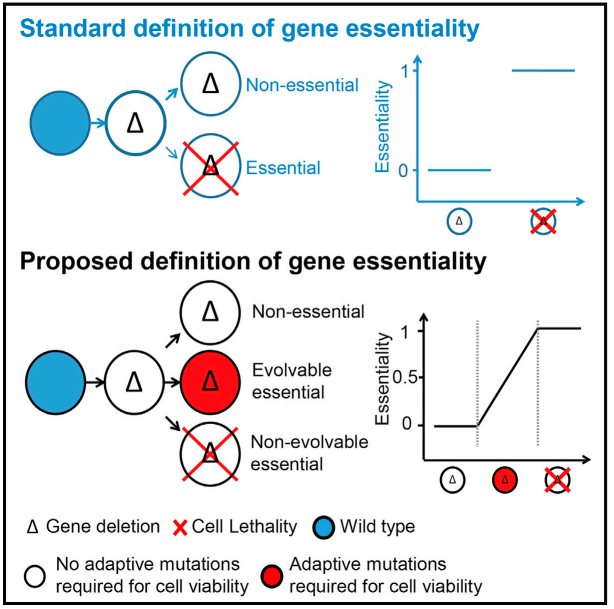
We screened ~1000 reported essential gene and found ~9% of them can be deleted from the genome without leading to lethality of the cell. We renamed these 88 genes as evolvable genes to distinguish them from non-essential genes and non-evolvable essential genes. Our finding revealed that gene essentiality is not a qualitative black/white trait but a quantitative trait linked to cellular evolvability.
Aneuploidy suggested adaptive mechanism
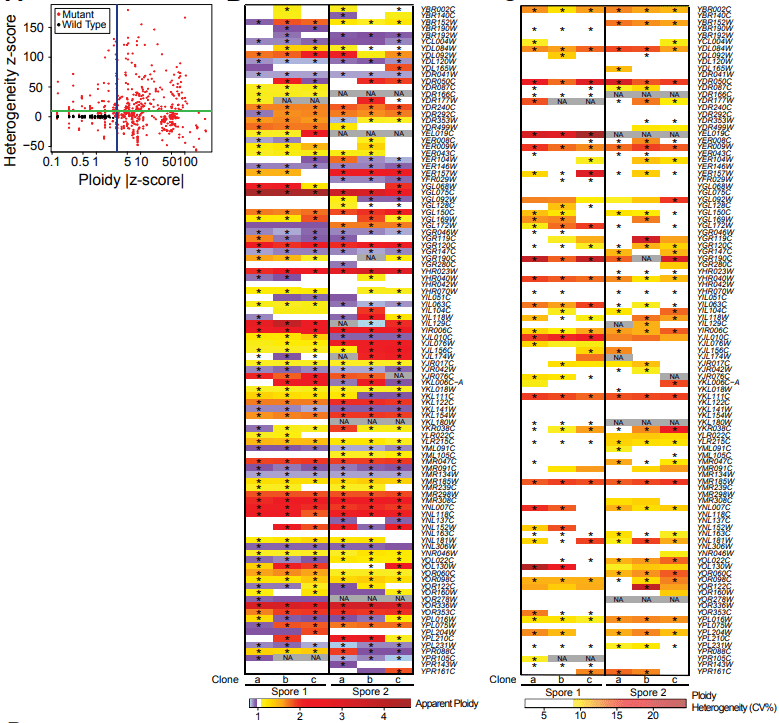
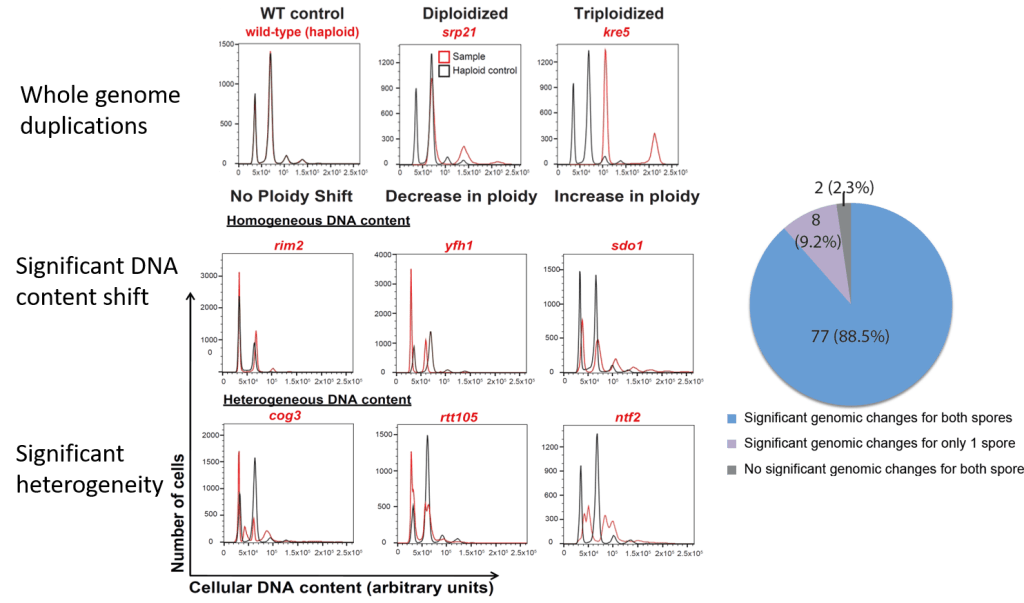
We analyzed more than 600 strains carried an evolvable gene deletion and discovered that the vast majority are aneuploid. These cells displayed different kinds of large scale genomic changes: whole genome duplication, disomy in haploids, monosomy in diploids, heterogeneous genome composition, etc. These observation suggested that these cells survived extreme stress- “essential gene deletion”- through aneuploidy.
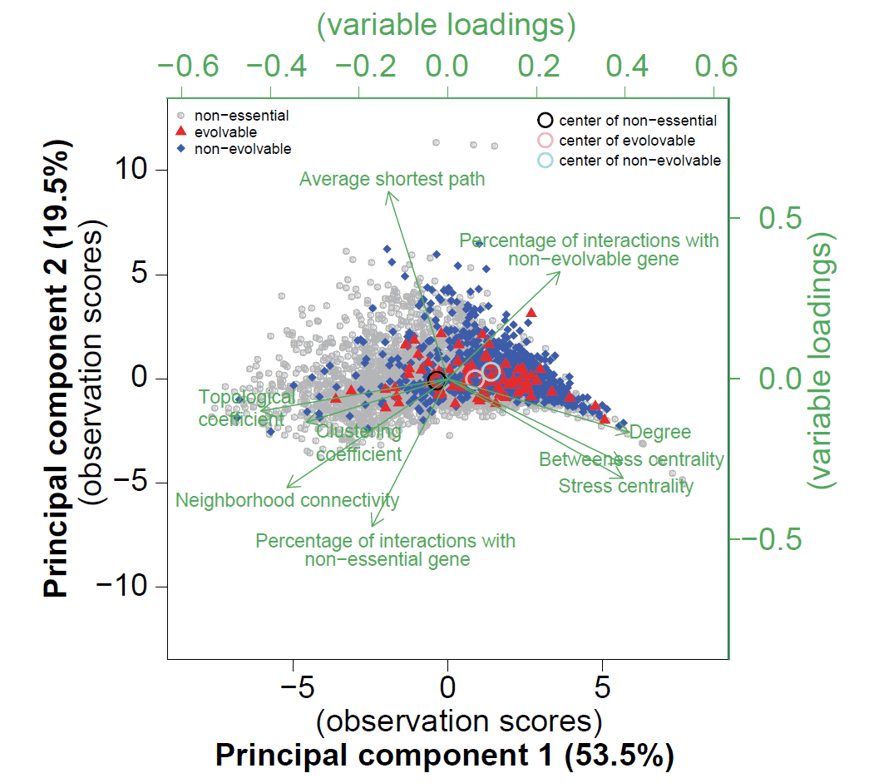
Evolvable genes form a continuum in between non-evolvable essential genes and non-evolvable genes.
We analyzed the conservation score/ network topology properties of the three class of genes: non-essential genes, evolvable genes, non-evolvable essential genes. We found that evolvable genes always positioned in between the other two classes.
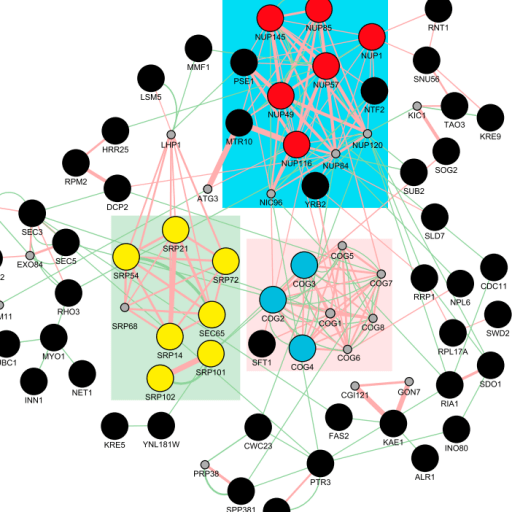
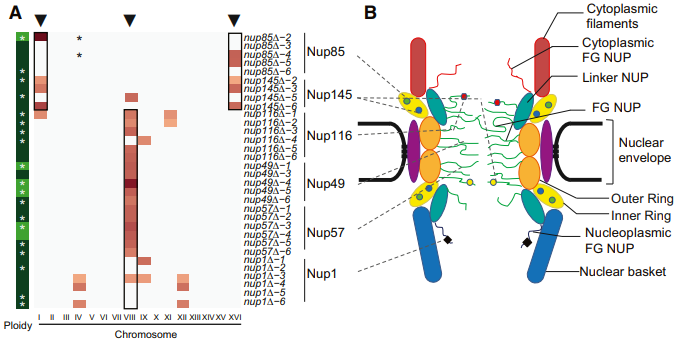
Evolvable genes were enriched in protein complexes involved in intracellular trafficking. Interestingly independently generated mutant strains carry the same gene deletion or deletion of genes involved in the
same protein complex carried specific set of aneuploidies, suggesting that aneuploidy could
function as an adaptive mutations and that cells respond to inactivation of different component of the same functional submodule in similar ways. Accordingly we showed that the presence of either specific aneuploidies or a specific gene on the aneuploid chromosome was required and sufficient to bypass the lethality of genes belonging to the same functional module.